Oct 31 2022
Alternative Gene Splicing – Another Method of Bioengineering
 Genetic engineering is a rapidly progressing scientific discipline, with tremendous current application and future potential. It’s a bit dizzying for a science communicator who is not directly involved in genetics research to keep up. I do have some graduate level training in genetics so at least I understand the language enough to try to translate the latest research for a general audience.
Genetic engineering is a rapidly progressing scientific discipline, with tremendous current application and future potential. It’s a bit dizzying for a science communicator who is not directly involved in genetics research to keep up. I do have some graduate level training in genetics so at least I understand the language enough to try to translate the latest research for a general audience.
Many readers have by now heard of CRISPR – a powerful method of altering or silencing genes that brings down the cost and complexity so that almost any genetics lab can use this technique. CRISPR is actually just the latest of several powerful gene-altering techniques, such as TALEN. CRISPR is essentially a way to target a specific sequence of the DNA, and then deliver a package which does something, like splice the DNA. But you also need to target the correct cells. In a petri dish, this is simple. But in living organism, this is a huge challenge. We have developed several viral vectors that can be targeted to specific cell types in order to deliver the CRIPR (or TALEN), which then targets the specific DNA.
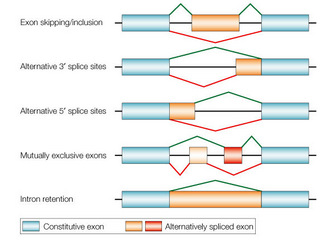
Now I would like to present a different technique I have not previously written about here – alternative splicing. A recent study presents what seems like a significant advance in this technology, so it’s a good time to review it. “Alternative splicing” refers to a natural phenomenon of genetics. Genes are composed of introns and exons. I always thought the nomenclature was counterintuitive, but the exons are actually the part of the gene that gets expressed into a protein. The introns are the part that is not expressed, so they are cut out of the gene when it is being converted into mRNA, and the exons are stitched together to form the sequence that is translated into a protein. Alternative splicing refers to the fact that the way in which the introns are removed and the exons stitched together can vary, creating alternative forms of the resulting protein. This dramatically increases the number of different proteins that an organism’s genes can code for, because each gene can potentially code for multiple protein variants through alternative splicing.
Alternative splicing can also be exploited as a method of controlling gene expression in a cell. This can be used for research and potentially for therapeutic purposes. This is now being used along with another method – the delivery of exogenous (from the outside) genetic material into specific cells and then coaxing the cell’s protein-making machinery to make proteins from the introduced gene. This is the same basic concept as the mRNA vaccines, which introduced mRNA of spike proteins from the COVID coronavirus into muscle cells, which then made the proteins to provoke an immune response.
The method used in the current study introduces a “construct” into cells – these constructs are either viral or plasmid, but essentially they contain a gene that you want a specific populations of cells to express into protein. As with CRISPR, however, you need to target the desired cell type, so that the new protein is not made everywhere, causing unwanted side effects. Several methods are used to accomplish this targeting. Some use features on the surface of specific cells in order to target them with viral or plasmid proteins. Viruses normally do this anyway, which is why a viral infection will affect certain cell types and not others. Also researchers can include microRNA in the construct that is being introduced into cells that will inhibit expression in unwanted cells. These techniques collectively are about 5% effective, which means there is a lot of off-target expression.
The Johns Hopkins researchers of the current study developed a technique to exploit alternative splicing as a way of targeting specific cell types. This is because patterns of alternative splicing can be happening in some cell types and not others. In fact, this is often how liver cells, for example, differ from spleen cells – they express different genes, but also have different splicing patterns. In their study they used a viral vector (Adeno-associated virus – AAV) because it is a safe and effective vector. However, AAV has a size limit, and many gene constructs are far larger than can fit into an AAV vector.
What they did is utilize the fact that now have vast databased of genes, their expression, and their alternative splicing across many species. We also have the computer power to crunch through these massive datasets and find potential alternative splicing sequences that are small enough to fit inside an AAV. Doing so they were able to make a construct that targeted a specific cell type, not only by the traditional methods, but also by targeting specific alternative splicing patterns. With this technique they were able to get their specificity up from 5% to 50%.
This is dramatically better, but still there is a lot of room for improvement. The question now is – is this 50% good enough for therapeutic purposes? It’s plenty good for research, and that in itself is incredibly useful. But is that still too much off-target expression that side effects will be too great? I suspect it depends on the intended application.
One potential application of this technology is to replace proteins that are missing from a specific cell population because of a genetic mutation. Some genetic diseases result from such missing proteins, while others may result from a dysfunctional or even toxic mutant protein. For those that are simply missing (the protein is not made at all, or only an extremely truncated and non-functional portion), simply introducing the gene and then forcing its expression can be tantamount to a cure. If the same protein is made in off-target cells, there may be side effects, but depending on the protein and its effects, these may be minimal.
Another application is to target cancer cells and kill them. If a specific type of cancer involves a specific alternative splicing pattern, it can be targeted, and a construct containing the gene for a protein that will essentially kill the cell can be introduced. Of course, off target effects are likely to be much more severe for such an application.
This is a good representation of the feedback loop that is currently at work within genetics. As our knowledge of genetics improves, this gives us more powerful tools to study genetics itself, which increases our knowledge and the power of our tools, and the cycle continues. This study is just one step in that process.






