Jun 13 2017
The CRISPR Controversy
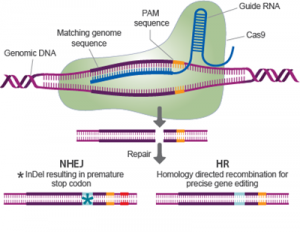 I suspect that CRISPR is rapidly following the path of DNA in that many people know the abbreviation and what it refers to but not what the letters stand for. Clustered regularly interspaced short palindromic repeats (CRISPR) is a recently developed technology for making precise gene edits. Such technology carries a great deal of promise for treating or even curing disease, for accelerating research, and for genetic modification technology.
I suspect that CRISPR is rapidly following the path of DNA in that many people know the abbreviation and what it refers to but not what the letters stand for. Clustered regularly interspaced short palindromic repeats (CRISPR) is a recently developed technology for making precise gene edits. Such technology carries a great deal of promise for treating or even curing disease, for accelerating research, and for genetic modification technology.
However a recent study has thrown some water on the enthusiasm for CRISPR and sparked a mini-controversy. The authors looked at two mice that were treated for blindness with CRISPR-cas9, sequencing their entire genome. They found over 1,500 unintended mutations. That would be bad news for the technology, which is revolutionary partly because it is supposed to be so precise.
For a little background, CRISPR was discovered in bacteria and archea. It is essentially part of their immune system – locating inserted bits of DNA from viruses and clipping them out. Researchers realized they could use this system to target specific sections of a genome to insert or remove a genetic information. The technique is fast, cheap, and convenient.
What this has meant is that genetic modification can now be cheaply available to even small labs. Further, since the technique can be used in living organisms, it could theoretically be used to cure genetic diseases.
Researchers already knew that CRISPR can also create mutations in parts of the genome that have some similarity to the target region, so-called “off target” mutations. So they routinely will monitor at-risk parts of the genome to detect such off target mutations. This new study, however, sequenced the entire genome, looking for off-target mutations even where they weren’t expected. That is why, the researchers believe, they found so many more off-target mutations than prior studies. They warn that we must monitor more that just the high risk parts of the genome for such unintended mutations.
If the results of this study are true, that would certainly put a dampener on the enthusiasm for CRISPR, especially as a therapeutic tool (not as much for research).
However, there has already been a backlash from the research community, many of whom are criticizing the study for methodological flaws and overcalling the results. For example, geneticist Eric Topol told Gizmodo:
“I think the Nature Methods paper was a false alarm on CRISPR induced mutations. Ironically, the methods used were flawed. While we remain aware of such concerns —unintended genomic effects that might occur with editing—that report was off-base.”
Essentially critics are saying that the researchers counted mutations incorrectly. They did not do a before and after comparison, but rather compared the CRISPR treated mice to other mice in their litter. However, some argue that those mutations can simply be due to natural variation. The researchers did not prove they were due to CRISPR.
What this study has done is raise a concern, but not really prove that there is a problem with CRISPR. What needs to happen from here is that this preliminary study be followed up with a more rigorous study that will address all the criticisms. It seems like the needed study should have more CRISPR treated animals and do before and after genome sequencing. That would likely settle the debate.
Once we know what the real risk of unintended mutations from CRISPR is researchers can then address the issue. Perhaps the technique needs to be modified (if possible). In any case, I don’t think concerns off off-target mutations are going to kill CRISPR. It is still clearly a revolutionary research tool. It may be more complicated to use as a therapeutic tool than previously assumed, but I doubt that will be an unsurmountable problem.
It is, as I often point out, difficult to predict technological advance, partly because of unexpected hurdles like this. In the 1990s using retroviruses to deliver gene therapy was a promising technology, but this was stalled because of unintended effects from the viruses, specifically resulting in leukemia in some patients. The technology is still useful for research, but as a therapeutic tool these safety concerns have essentially derailed it.
It remains to be seen if CRISPR will go the same route or will overcome these apparent safety hurdles.






