Nov 20 2018
Possible New Branch of Eukaryotes Defined
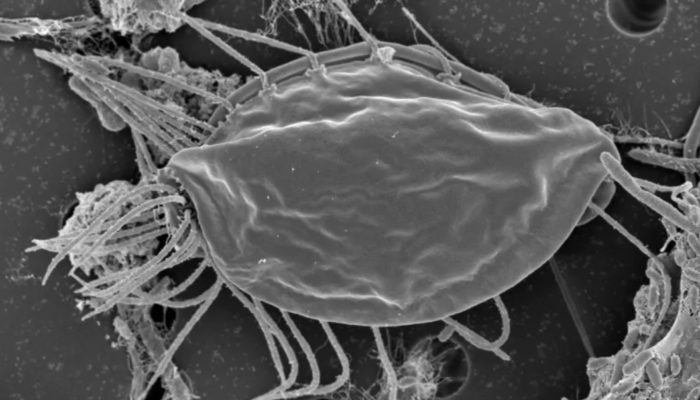 Scientists report in Nature the indentification of two new species of Hemimastigophora, a predatory protist. What makes the paper newsworthy is that the authors are arguing that their genetic analysis suggests Hemimastigophora, currently categorized as a phylum, should instead be its own suprakingdom.
Scientists report in Nature the indentification of two new species of Hemimastigophora, a predatory protist. What makes the paper newsworthy is that the authors are arguing that their genetic analysis suggests Hemimastigophora, currently categorized as a phylum, should instead be its own suprakingdom.
To make sense of this let’s review the basic structure of taxonomy, the system we use to categorize all life. All known life is divided first into three domains, the bacteria, archaea, and eukaryotes. Bacteria and archaea do not have a nucleus, while eukaryotes are larger and have a nucleus which contains most of their DNA.
Eukaryotes are divided into kingdoms, including plants, animals, fungus, protozoa, and chromista (algae with a certain kind of chlorophyll). Kingdoms are then divided into phyla, which are essentially major body plans within that group.
This is a simplified overview, because there is a lot of complexity here, with suprakingdoms, subkingdoms, and further breakdowns. Further, there is a lack of consensus on how to exactly divide up these major groups. Even in the cited paper, the authors say there are 5-8 “suprakingdom level groups” within the domain eukaryotes. The number of kingdoms depends on which scheme you use, and how you interpret the existing evidence.
The reason for uncertainty is that we have not yet done a full genetic analysis on every known group. Further, when we discover new species that lie outside of the existing scheme, we have to rethink how different groups are actually related.
The Hemimastigophora were actually discovered in the 19th century, with 10 known species. The new paper presents the discovery of two new species – but again, the group has been known for over a century. They also present the first genetic analysis. They argue that their analysis supports elevating the group from a phylum to a suprakingdom. That would be a huge change.
Specifically they state that the Hemimastigophora are more different from animals and fungi than either group is from each other. This is not as weird as it sounds, because at that level animals and fungi are actually “closely” related. They are in the same supergroup, the Opisthokonta, which contains animals, fungi, and choanoflagellates (a single-celled eukaryote).
This also highlights how genetic analysis is changing classification systems. We cannot simply go by how organism look. That is the old way, which would put, for example, all single-celled eukaryotes into one group. But at the molecular level there is a lot of variation among the single-celled eukaryotes, one branch of which went on to evolve into animals, while another evolved into plants, for example. When we separate them at the molecular level, reflecting evolutionary relationships, then groups which look very different (animals and fungus) end up together.
Obviously we are talking about deep evolution, 500 million – 1 billion years ago or more. These are evolutionary divisions that were occurring while the Earth was populated only by single-celled organisms. Only by genetic analysis can we hope to untangle their complex relationships.
So really what’s new about this paper is that it is the first recategorization of Hemimastigophora based on genetic analysis. Putting it in its own more fundamental group will help scientists think about these early evolutionary relationships.
I don’t know how long it will take for biologists to arrive at a consensus for the exact relationships at this level. It has largely to do with grades vs clades. Grade is a proposed term for a group that is based on morphology (how critters look) rather than genetics, while a clade is an evolutionary group. Bacteria, for example, are really a grade, not a clade, and are therefore likely polyphyletic.
A monophyletic group is a group with a single common evolutionary ancestor. A polyphyletic group contains multiple evolutionary branches, and lack a single common ancestor.
So my desire for a neat tidy system I can easily memorize is frustrated by messy reality. Evolution is messy at every level, because branching can occur at any point, with various kinds of changes. Further, traits can be acquired, then later lost. To make matters worse, there is horizontal gene transfer, where genes from one group (even kingdom) can contaminate another (usually through viral transfer).
Part of the messiness also comes from outliers. Even if one species survives from an early branching, we need to adjust the classification system to accommodate it (the duck-billed platypus effect). So we end up with suprakingdoms, subphyla, and really no one clean system that everyone can agree on.
But I guess we have to embrace the messiness. It is a fundamental feature of life, and we shouldn’t try to force it into our desires for a neat system.
In any case – it isn’t every day we add a new branch of life above the kingdom level. But this is also probably not the last time this will happen either. We are in the middle (taking the long view) of a massive reclassification of all life based upon genetic analysis. Things will probably settle down once we get mostly to the other end of this process.






