Jul 06 2017
The Brain’s Wiring
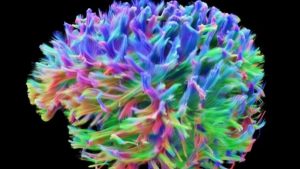 Cardiff University has released its latest scans of the wiring of the human brain. This now adds to a similar project in the US funded by the NIH.
Cardiff University has released its latest scans of the wiring of the human brain. This now adds to a similar project in the US funded by the NIH.
The result is a stunning image of all the axons in the human brain – the wires that conduct signals and form networks and connections to other parts of the body. In addition to these wires there are also the neurons, which are the cell bodies of which the axons are part, and the glia, which are other cells in the brain that serve support functions but also play a role in modulating neuronal function.
These wiring images are part of various connectome projects – attempts at fully mapping all the connections within the human brain and their functions. These latest images from Cardiff are the result of the Siemens 3 Tesla Connectome MRI system. Essentially we are seeing the result of advances in both hardware and software technology.
The MRI scanners themselves are on the more powerful side. The “3 Tesla” refers to the power of the magnet. A typical hospital MRI scanner operates at 1.5 Tesla or 3 Tesla. There is a 10.5 Tesla MRI unit at the University of Minnesota which is used for the NIH connectome project. So 3 Tesla is a powerful MRI scanner, but no more so than the more powerful typical clinical MRI scanners.
The power of the magnet, however, is not the only measure of the abilities of an MRI scanner. The Cardiff unit is “specially adapted” to have very high resolution. They can image fibers in the brain 1/50th the width of a human hair. There is only one other unit with this resolution in the world, at Harvard University.
In addition to high-resolution anatomical imaging, the Cardiff and NIH projects use function MRI scanners to image the axons in action. This information is supplemented with magnetoencephalography (MEG) and EEG scanning, which are also functional scans looking at brain activity. The goal is to combine anatomical and functional data to create the connectome – a map of the functional networks within the brain.
The resulting images are stunning. It reminds me of the Saturn V rocket in a way. In response to the moon hoax nonsense, one NASA engineer commented that – looking at the Saturn V rocket’s size, and doing even some basic calculations, it’s clear that rocket was designed to go beyond Earth orbit. It was large enough not only to reach the Moon, but theoretically even Mars. If we didn’t really go to the moon, why was the rocket so big?
I feel the same way looking at the emerging images of the connectome. If you look at all the connections in the brain, you have to wonder what they are for. All those networks and all that processing power is needed to do all the things that the brain does, including producing consciousness. I know that won’t convince neuroscience deniers, but for scientists it is amazing.
One comment on reporting about these images. The BBC writes:
Doctors hope it will help increase understanding of a range of neurological disorders and could be used instead of invasive biopsies.
and
The scanner is being used for research into many neurological conditions including MS, schizophrenia, dementia and epilepsy.
I know it is obligatory for any basic science reporting in the media to make a connection to some concrete application. It is, of course, a reasonable question to ask – how can we exploit this new technology or finding for something specific? Unfortunately, that is often where speculation goes off the rails, or at least is not put into perspective.
I do have great hopes for the potential of the connectome project, but it won’t lead directly to cures for MS (multiple sclerosis) or dementia, and it may not even contribute to their diagnosis. First let’s address research into understanding these diseases.
Some brain disorders, like MS and Alzheimer’s disease (AD), are pathological diseases. MS is caused by inflammation in the brain and spinal cord which causes direct damage to cells and axons. In AD cells in the brain slowly die off for still unclear reasons, although we have identified many pathological changes that are involved in the progression of the disease.
In other words, these are pathological diseases of the cells in the brain. They are not the result of disordered connections within the brain. Brain connections are disrupted secondary to the pathology, so they are the result of and not the cause of the disease.
This kind of imaging, therefore, might help us understand the consequences of diseases like MS and AD. I don’t think, however, they will help us understand the pathology, and therefore is unlikely to lead to effective treatments (which have to address the underlying pathology).
Schizophrenia is different. That is a disorder of brain wiring, and not necessarily pathological. Brain cells are fine, they are just networked to each other in a way that produces the signs and symptoms we define as schizophrenia. Connectome projects, therefore, will likely be much more useful to understanding disorders like schizophrenia which are the result of disordered connections.
What about using this technology as a diagnostic tool. That is possible, but not as straightforward as reporting makes it seem. In order to be useful clinically, any examination tool must give us specific clinical information that we can act upon. It has to distinguish different related diagnoses, or make a diagnosis earlier than we can by other methods, or predict outcome or who will respond to specific treatments. Just knowing in more detail exactly how a brain is misfiring won’t necessarily lead to any change in management.
I do not think it is likely that connectome scans will be useful in diseases like MS. I don’t see how this information will affect management. Further, I don’t think they will replace biopsies (to be clear, we don’t routinely use biopsies to diagnosis MS). Biopsies are about finding pathology, and as I said above, imaging the connections is not about cell pathology. They are simply different kinds of information.
The only clinical role I can envision for AD is making a diagnosis earlier than can be made clinically. Sometimes patients present with very mild symptoms of dementia, before they are advanced enough to confirm the diagnosis of AD or a similar dementia. Such a scan might show early connection loss and confirm the diagnosis a year or so before it can otherwise be confirmed. I don’t know if this will be the case, but it’s plausible.
Whether or not this is clinically useful is a separate question. It will be helpful for prognosis, which will help people plan for their future. It may not otherwise have a dramatic affect on management, but that may change as new treatments become available.
Overall I think the direct clinical applications are likely to be limited, but may find utility in specific situations. The research applications, however, may be massive, but they will not apply to pathological disease. Rather they will mostly apply to our understanding of how the healthy brain works, and in understanding disorders which are essentially problems with the brain’s connections.






